Coronavirus disease 2019 (COVID-19), an acute respiratory distress syndrome caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is currently a global public health concern. As of August 28, the world has reported more than 24 million confirmed cases, and the death toll has scaled 800,000.For coronavirus, the nonstructural protein NSP is of great significance. “CoV nonstructural proteins (Nsps) play pivotal roles in the assembly of this complex and associated enzymatic functions in virus genomic replication.”(Abstract) The research team of Professor Dan Su from the State Key Laboratory of Biotherapy, Sichuan University has published in Molecular Biomedicine a research paper entitled "Structural Basis for the Multiplication of Nonstructural Protein Nsp9 from SARs-CoV-2", which analyzes in depth the structure of Nsp9, the nonstructural protein of new coronavirus. Professor Su is the corresponding author and the first author is Changhui Zhang of Sichuan University.
“The genome of SARS-CoVs consists of an approximately 30-kb non-segmented positive-sense RNA sequence with a 5′ untranslated region (UTR), followed by a single open reading frame and a short flanking 3′ UTR [9]. The life cycle of SARS-CoVs starts with its binding to the cell surface receptor angiotensin-converting enzyme 2 (ACE2) through its spike glycoproteins, resulting in receptor-mediated endocytosis [10]. Upon entry into the host cell, the virus expresses its replicase gene to initially generate two precursor polyproteins, pp1a and pp1ab. These then undergo cleavage to yield 16 mature nonstructural proteins (nsp1–16) as well as several intermediate precursors by two distinct viral proteinases, the papain-like proteinase within nsp3 and a 3C-like proteinase nsp5 [11].”(Introduction)
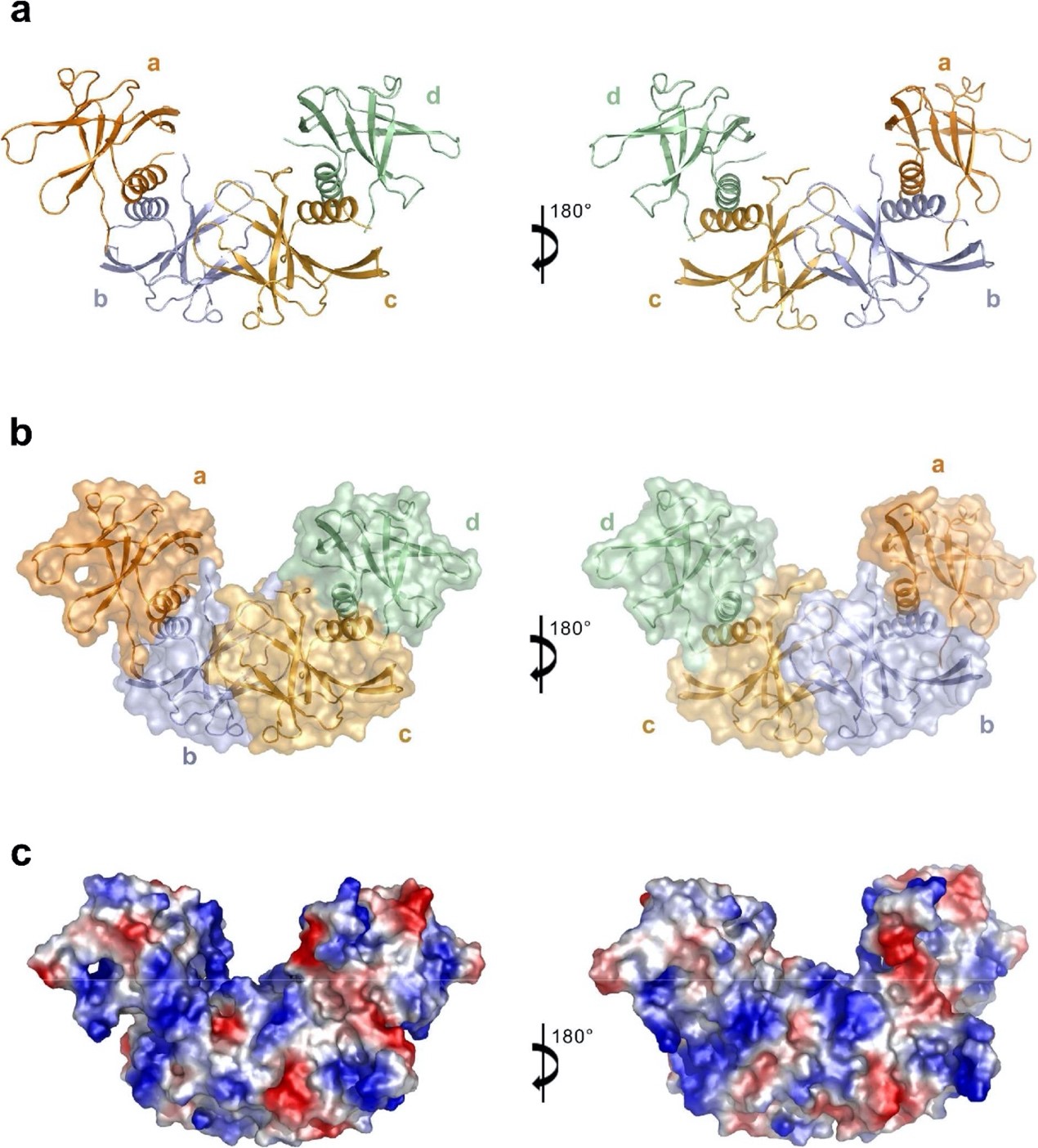
Fig. 1 Structure of the SARS-CoV-2 nsp9 tetramer.
The two stable binding surfaces in the tetramer are closely related to the binding ability of coronavirus to nucleic acid. Therefore, the author observed the nucleic acid binding ability of SARS-CoV-2 nsp9 by EMSA. This experiment confirmed that SARS-CoV-2 nsp9 has binding ability to ssRNA and ssDNA of different length, and has certain preference: sars-cov-2 Nsp9 has stronger binding ability to long strand RNA (Fig. 2).
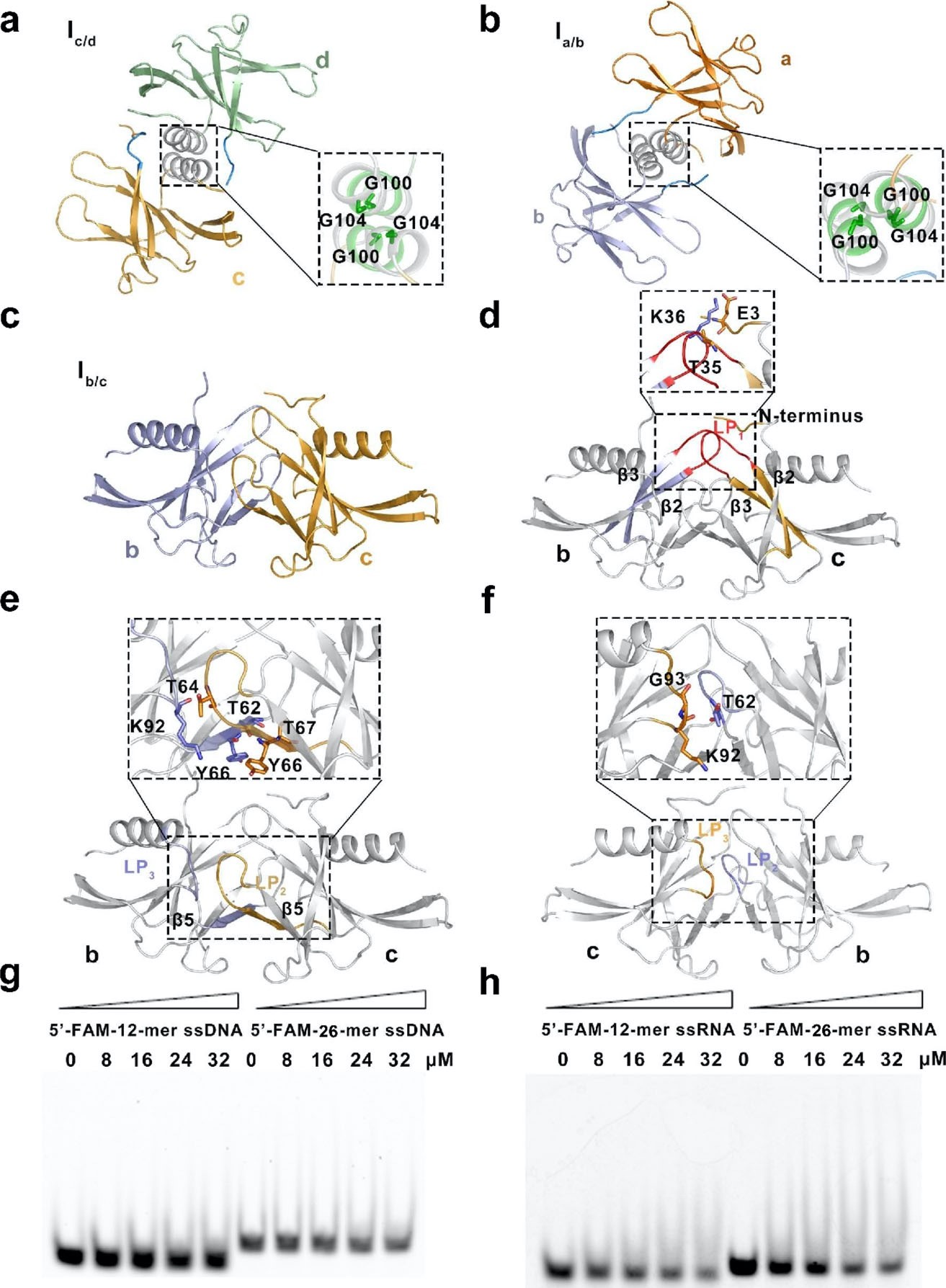
Fig. 2 Details of structure of the SARS-CoV-2 nsp9 tetramer and function in nucleic acid binding.
”In this study, we determined the structure of SARS-CoV-2 nonstructural protein 9 (nsp9), an RNA-binding protein that is essential for CoV replication. Its homotetrameric structure with two stable dimeric interfaces provides a structural basis for understanding the mechanisms of RNA-binding protein self-assembly, which may be essential for the regulation of viral RNA replication and transcription.” (Abstract)
Article link: https://doi.org/10.1186/s43556-020-00005-0
