The research team of professors Weimin Li and Dan Xie at West China Hospital has recently published their discovery on non-small cell lung carcinoma in Cancer Research ( IF: 8.378 ). Being established as the first cancer periodical, Cancer Research of American Association for Cancer Research ( AACR ) is the largest prestigious academic publication in the world. SCU West China Hospital has conducted this research independently. Professor Weimin Li of Pulmonary and Critical Care Medicine ( PCCM ), West China Hospital and Professor Dan Xie of SCU State Key Laboratory of Biotherapy are the corresponding authors. The research has been funded by National Natural Science Foundation of China.

Figure 1 Title
Lung cancer is one of the leading causes of cancer-related death worldwide. Due to its complex pathogenesis, there is no effective cure so far. In recent years, genome sequencing of lung cancer has revealed the association between some gene mutations and lung cancer. However, there are still some patients with lung cancer who do not have obvious driver gene mutations, but have changed at the epigenetic level. Among them, the open region of chromatin is an important epigenetic feature. The presence of open chromatin often means the activation of corresponding DNA regulatory elements to regulate downstream gene expression. In light of the complex gene regulation model, it is impossible to analyze the process of tumorigenesis and development by observing only one dimension of data changes. Therefore, it is of great significance to integrate multiple sets of data ( genome, repository, epigenetic group ) to reveal the complex gene regulatory network of lung cancer from multiple levels. The team integrated ATAC-seq, genome-wide sequencing and transcriptome sequencing techniques to study gene regulation in lung cancer. Through the analysis of chromatin open regions in 50 cases of non-small cell lung cancer, the researchers found that different pathological types of lung cancer have their own characteristics of open regions: For example, the specific open region of lung squamous cell carcinoma appears around genes related to the process of epithelial keratinization, and the specific open region of early lung adenocarcinoma appears around genes related to prognosis, etc.

Figure 2 Research Design
In addition, the researchers found wider open chromatin peaks in lung cancer samples. These special open regions are between 1kb and 180 kb in length, mainly enriched near the transcriptional initiation sites of genes. They not only show the unique distribution characteristics of cancer species, but also are related to the degree of confusion in gene expression. The expression of lung cancer driving genes such as EGFR and ERBB3 in these regions fluctuated greatly in the samples. Combined with the data of chromosome structure variation in DNA sequencing, the team also found that wide open regions were enriched in gene fragments with increased genomic copy numbers. At the same time, genome copy number variation (CNV) was also strongly associated with signal and gene expression in open regions, i.e. CNV would affect gene expression regulation in coordination with chromosome open regions. In this study, the team observed a large number of genes affected by this expression pattern.
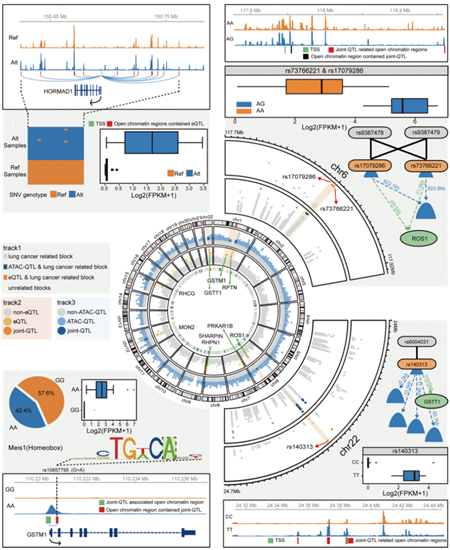
Figure 3. Molecular regulatory network of lung cancer based on multigroup data
Multigroup data provide convenience for the analysis of gene regulation patterns in lung cancer. At the end of the paper, the research team revealed the complex molecular regulatory network between genome sequence variation, large fragment DNA structure change, open chromosome region and gene expression change by analyzing the correlation between different histological information.
In this study, the genome regulatory network of non-small cell lung cancer ( NSCLC ) was revealed by integrating multi-genomic data. Advanced genomics methods were used to identify different subtypes of lung cancer. Twenty-one quantitative trait loci ( joint-QTLs ) related to open genome regions and transcript expression regulation were identified. It further confirmed the model of GWAS study that the sites associated with lung cancer risk participate in the genome regulatory network.These regulatory sites located in non-coding regions could be used as targets for lung cancer diagnosis and drug design to guide the precise treatment of lung cancer.
The article link: https://cancerres.aacrjournals.org/content/early/2019/08/08/0008-5472.CAN-18-3663.full-text.pdf
