Mature oocytes of mammals have the ability to reprogram sperm and somatic nuclei to produce new offspring or cloned individuals. During the maturation of mammalian oocytes, its epigenetic modification (such as DNA methylation) and chromatin status also change dramatically. However, the relationship between chromatin remodeling characteristics and DNA methylation and chromatin status in this process is not very clear.
On December 18, Fan Guo Team published their latest research results online in Cell Research (IF:15.4), entitled Integrative single-cell analysis of transcriptome, DNA methylome and chromatin accessibility in mouse oocytes. Researchers developed an improved single-cell COOL-seq technique (iscCOOL-seq) combined with single-cell transcriptome sequencing technology, bioinformatics analysis and vitro culture system of mouse follicles to analyze a total of 1,394 oocytes and follicles in mice. They draw the transcriptome maps, DNA methylation maps and chromatin status maps during oocytes maturation at the resolution of single cell and single base and analysis heterogeneity at different histological levels and multihistology. At the same time, the key molecular signaling pathways during mouse oocytes maturation were validated by vitro culture system of mouse follicles.

Researchers first invented a new method to construct single-cell DNA methylation library, TAILS, and systematically improved the existing single-cell COOL-seq technology by using this method, making the new improved iscCOOL-seq multi-omics sequencing technology retain high accuracy and accuracy, while significantly improving the comparison rate of sequencing reading segments. Subsequently, 1030 mouse oocytes were analyzed in several representative stages of oocytes development, and the following new findings were made:
(1) In the process of oocytes growth and maturation, single oocytes undergoes drastic changes in chromatin state before and after the initiation of growth, and the transcription group also go through significant changes accordingly; however, the overall DNA methylation level of the genome is still below 5% before and after the initiation of oocytes growth, and there is no significant increase (figure below),which suggested that non-growing oocytes have unique gene ex-x-pression and chromatin accessibility.
A diagram of the number and diameters of growing oocytes used at each stage.
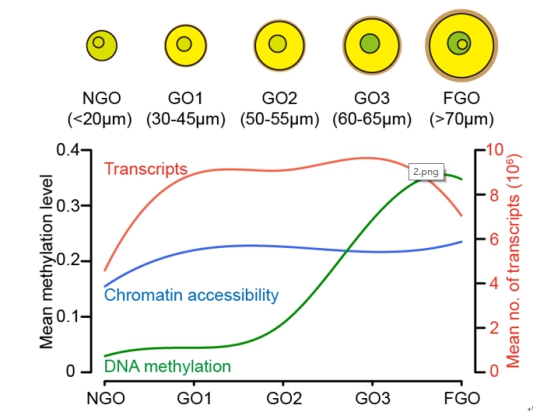
(2) In the process of oocytes growth and maturation, the heterogeneity of DNA methylation among individual oocytes was significantly enhanced, especially in the repetitive elements and heterochromatin regions of the genome; the heterogeneity of chromatin state was higher in the open chromatin regions.
(3) In the process of oocytes growth and maturation, open chromatin is mainly concentrated in promoter, CpG island and H3K4me3 modified chromatin, which often have low DNA methylation level. For Partial Methylated Domains (PMDs) in genome, the enrichment of open chromatin is much lower than that in other regions of genome, which suggested that DNA methylation level is closely related to the degree of open chromatin enrichment during the growth of oocytes.
(4) During the growth and maturation of oocytes, the open chromatin was enriched near the transcription start site (TSS) and transcriptional end sites (TES), while the enriched open chromatin region near the TSS often overlapped with the gene alternative promoters. It is suggested that the gene promoter of oocytes has unique chromatin state characteristics.
(5) During the growth and maturation of oocytes, de novo DNA methylation often occurs in the Gene Bodies region. For these region, the promoter region of the corresponding gene is open as early as the non-growth oocytes stage; for the Gene Bodies region where initiative DNA methylation does not occur, the promoter region of the corresponding gene tends to be closed. It is suggested that there is a precise regulation relationship between the chromatin status of gene promoter, gene transcription and initial DNA methylation in oocytes.
(6) During the growth and maturation of oocytes, some of the epigenetic factors represented by histone modification-related factors will present stage-specific ex-x-pression; binding motif of Rbpj, which is a downstream TF of the Notch signalling pathway, showed significant enrichment. These results suggest that epigenetic and transcriptional regulation plays an important role in oocytes maturation.
Finally, the role of Notch signaling pathway, a key molecular signaling pathway identified by single cell multi-omics analysis, in oocytes maturation was validated by using small molecule inhibitors combined with vitro culture system of mouse follicles. By recording and culturing 364 mouse follicles in vitro, the researchers found that after treated with small molecule inhibitors targeted to Notch signalling pathway, the in vitro maturation of follicles was compromised, indicating an important role of Notch signalling during oocytes growth.
This study systematically depicted the precise and orderly changes of DNA methylation, chromatin status and gene ex-x-pression during the development and maturation of mammalian oocytes, as well as the relationships among various histological levels. It will lay a foundation for further research on the reprogramming ability of mammalian oocytes and provide a new idea for the improvement of somatic cell clone efficiency and the study of abnormal development of oocytes.
(The report was transferred and rearranged from WCSUH.)
