The correct allocation of chromosomes to daughter cells during mitosis depends on the assembly of kinetochore protein complexes on the chromosome centromere. The attachment of kinetocytes and spindle microtubules is strictly monitored by thespindle assembly checkpoint (SAC) to delay cell entry into the late stage until all kinetocytes are properly attached. Arabidopsis KNL1 is only related to animals in two functional domains, and the remaining amino acid sequences have almost no homology. Can plant homologs play a role in SAC signaling? Do highly differentiated KNL1 family proteins have different functions in different plant lineages? These issues are worth studying.
Honghui Lin/Xingguang Deng’s research team (College of Life Sciences, SCU), and Bo Liu’s team (the University of California, Davis) collaborated to publish a research paper titled "A Coadapted KNL1 and Spindle Assembly Checkpoint Axisorchestrates Precise Mitosis in Arabidopsis” in PNAS recently. The paper analyzes the rapid evolution and lineage specific connections between kinetochore protein KNL1 and spindle assembly checkpoint signaling molecules. Xingguang Deng, an associate research fellow and doctoral students Ying He and Xiaoya Tang from the School of Life Sciences, SCU are the co first authors of the paper. Prof. Honghui Lin, Xingguang Deng, and Prof. Bo Liu are the co corresponding authors of the paper. This research work was funded by the National Natural Science Foundation of China, the Sichuan Provincial Natural Science Foundation of China, the Sichuan University Innovation Team and the National Science Foundation of USA.

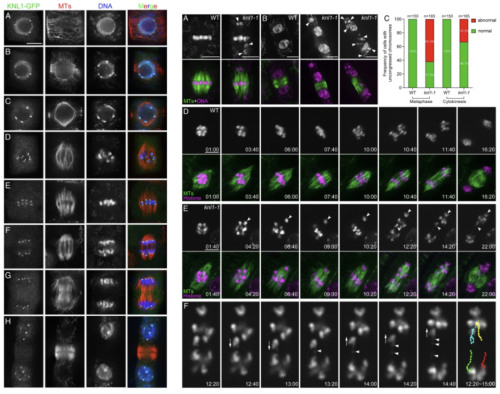
KNL1 Plays a Critical Role in Faithful Chromosome Segregation.
"The kinetochore scaffold 1 (KNL1) protein recruits spindle assembly checkpoint (SAC) proteins to ensure accurate chromosome segregation during mitosis. Despite such a conserved function among eukaryotic organisms, its molecular architectures have rapidly evolved so that the functional mode of plant KNL1 is largely unknown. To understand how SAC signaling is regulated at kinetochores, we characterized the function of the KNL1 gene in Arabidopsis thaliana. The KNL1 protein was detected at kinetochores throughout the mitotic cell cycle, and null knl1 mutants were viable and fertile but exhibited severe vegetative and reproductive defects. The mutant cells showed serious impairments of chromosome congression and segregation, that resulted in the formation of micronuclei. In the absence of KNL1, core SAC proteins were no longer detected at the kinetochores, and the SAC was not activated by unattached or misaligned chromosomes. Arabidopsis KNL1 interacted with SAC essential proteins BUB3.3 and BMF3 through specific regions that were not found in known KNL1 proteins of other species, and recruited them independently to kinetochores. Furthermore, we demonstrated that upon ectopic expression, the KNL1 homolog from the dicot tomato was able to functionally substitute KNL1 in A.thaliana, while others from the monocot rice or moss associated with kinetochores but were not functional, as reflected by sequence variations of the kinetochore proteins in different plant lineages. Our results brought insights into understanding the rapid evolution and lineage-specific connection between KNL1 and the SAC signaling molecules."(Abstract)

KNL1 deploys a eudicot-specific domain to recruit BUB3.3 and BMF3 to kinetochores.
https://doi.org/10.1073/pnas.2316583121
