Personalized precision medicine has become an emerging theme for biomedical research. Personalized precision medicine became well known in 2015 when the US President Barack Obama proposed the Precision Medicine Initiative. Due to the development of high-throughput technologies, we can systematically identify the association betweenpersonalized physiological states (such as disease onset and progression) and personalized molecular signature, thus achieving personalized monitoring, diagnosis and treatment.
Traditional large-scale cohort studies aim to understand the average states of a population with the same traits, such as disease or tumor, while ignoring the inter-individual differences and the individual’s specific changes. If we can monitor the molecular level data of an individual for a long time, then we will be able to discover the changes and predict diseases or other physiological state changes based on the individual's own average states rather than the average measurements of the population.
DNA methylation is a key mechanism in gene expression regulation and is influenced by environmental factors and age. And it is often asymmetric ­– for example, allelic differentially methylated regions (aDMRs) are associated with the preferential silencing of one of the two parental gene copies. The spatial and temporal regulation of DNA methylation is important in understanding biology and disease. However, only a few spatial and temporal studies have been performed. Detailed dynamics and the relationship between the DNA methylome and transcriptome as well as their association with different physiological states and health/disease condition are poorly understood.
In the study “Longitudinal Personal DNA Methylome Dynamics in a Human with a Chronic Condition”, which was published in Nature Medicine on Nov. 5th2018, the collaboration team of Michael Snyder (Stanford University) and Dan Xie (the SCU State Key Laboratory of Biotherapy) has monitored one healthy individual’s DNA methylome and transcriptome for three years, finding that dynamic alterations of the DNA methylome correlated with spikes in blood glucose levels, whereas transcriptome changed frequently with viral infections.

Figure 1: Title page of the article
All data collection and experimental content of this study were completed in Stanford University, and bioinformatics analysis was completed by the Dan Xie research team in Sichuan University. Xia (a Ph.d candidate) was in charge of the DNA methylation data analysis, and Tu (a Ph.d candidate) was in charge of the transcriptomic data analysis.
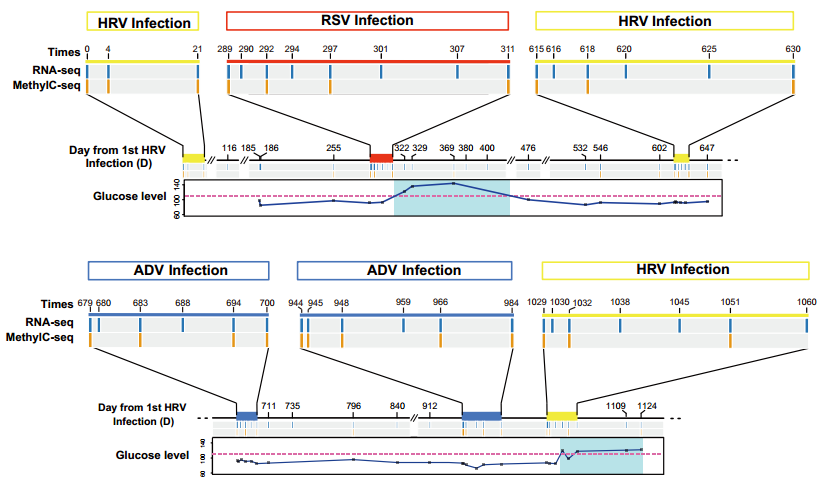
Figure2: Overview of sample collection and health condition.
The researchers collected peripheral blood mononuclear cells from a 54-year-old male volunteer at 57 times during a 36-month period and performed RNA-seq from all the time points and MethylC-seq from 28 time points for analysis. During this period, the volunteer experienced six viral infections and two elevated periods of fasting glucose levels and glycated hemoglobin A1c that reached diabetic levels.
In general, the researchers noted a high correlation among DNA methylation patterns of 28 time points. It suggested that the DNA methylation profiles were stable over time.Surprisingly, compared with other time points, the DNA methylation profiles of the time points during an adjacent elevated glucose level have higher similarity. When they focused on those events, they identified that the biggest number of differentially methylated regions (DMRs) arose between 80 days and 90 days prior to when the actual change in glucose occurred, and 476 DMRs were shared in these two glucose-elevated events.
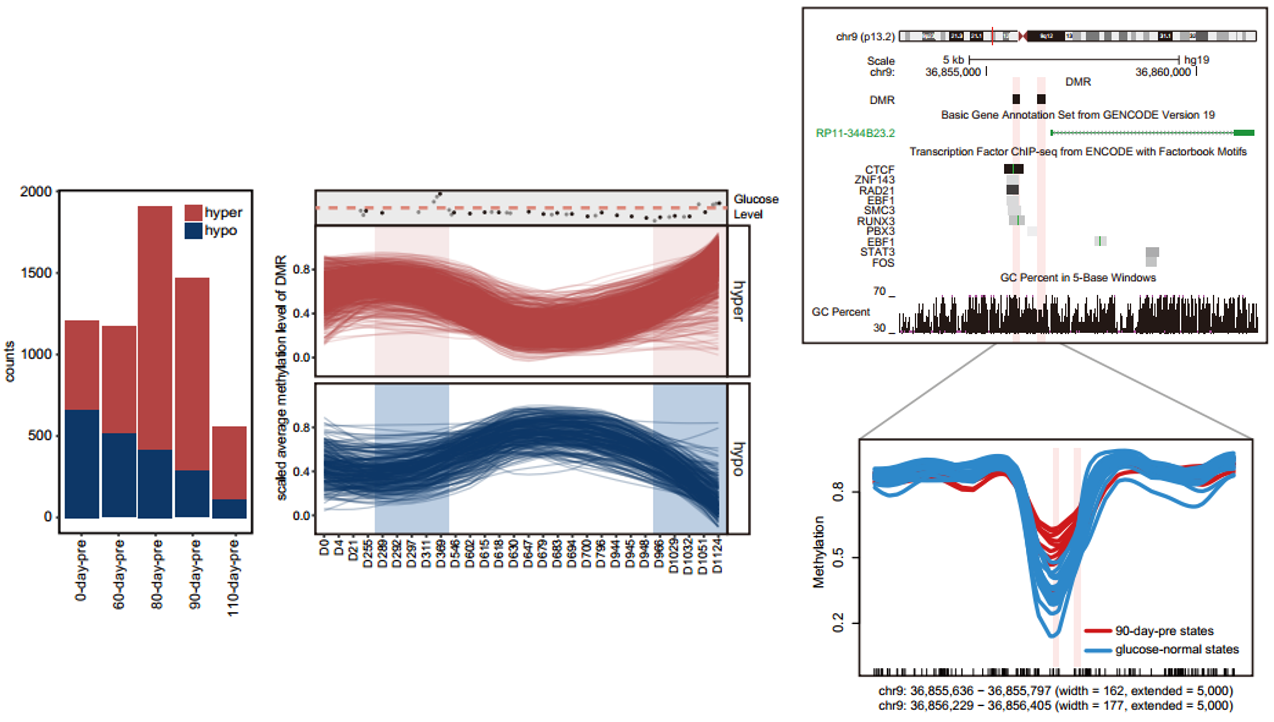
Figure 3: Differential methylation region between health and elevated blood glucose stages. Left panel shows the number of DMRs between health and multiple time periods before elevated blood glucose stage. Middle panel shows methylation level of two of DMRs appearing in both 80-day-pre and 90-day-pre time periods. Right panel shows genome browser view of methylation level around the two DMRs mentionedpreviously.
To further understand how longitudinal molecular profiles relate to health-related biological functions, researchers adopted a sliding window approach to investigating the short-term methylomic and transcriptomic changes. They found that there were more frequent short-term fluctuations in transcriptome data than methylome data. The short-term fluctuations of metylome data were associated with ‘glucose and diabetes’ related functional term/pathways, while the short-term changes of transcriptome data were associated with ‘immunologic process’ related functional term/pathways.
These results indicated that there might be a ‘phase-delayed alignment’ between longitudinal methylome changes and physiological states. In other words, since glucose level elevation is a chronic condition, the associated methylomic changes may occur before manifestation of the symptoms. The researchers found out that the higher percentage of differentially methylated genes in ‘glucose and diabetes’-related terms and pathways started at the 110-day-pre state, peaked at the 90-day-pre and 80-day-pre states, and dropped at the 60-day-pre state.
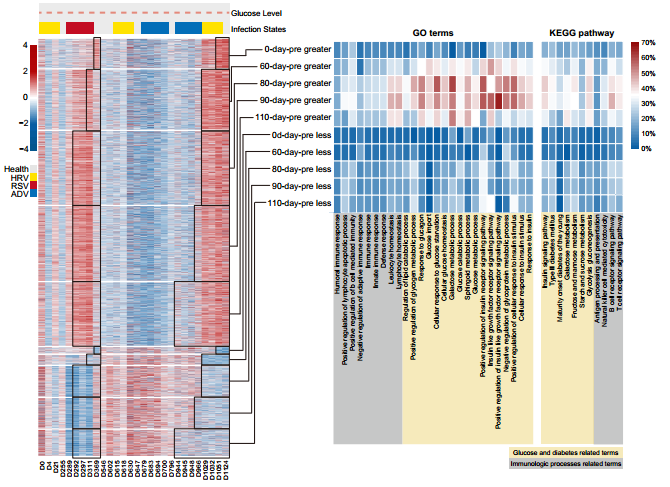
Figure 4: Methylomic changes were associated with glucose alterations.
To further explore potential differences of regulatory roles between differentially methylated sites associated with chronic conditions and those with acute conditions, the researchers defined cytosines differentially methylated between high-glucose-level and normal-glucose-level samples as glucose-dynamics-related differentially methylated sites (gDMSs), and cytosines differentially methylated between viral infection and non-viral infection samples as infection-related differentially methylated sites (iDMSs). They found that gDMSs were more enriched at promoter regions than iDMSs. That indicated the methylomic changes in promoter were morepronounced in chronic diseasesthan viral infection. Furthermore, they found 5 of the previously reported 149 markers overlapped with gDMSs.
The researchers also found that transcriptomic changes were linked to the viral infections. For instance, as compared with healthy time points, 116 genes were differentially expressed during two adenovirus infections, including 56 genes whose expression changed at specific time points during viral infection.

Figure 5: 116 DEGs shared by 2 ADV infection events. 56 genes whose expression changed at specific time points during viral infection were boxed out.
Finally, the researchers identified a much larger amount of allele-specific differentially methylated regions (aDMRs) than previous reported (~11-fold greater than previous studies). They also found the number of allele-specific DMRs is highly correlated with the number of genes on each chromosome. It suggests that aDMRs tend to co-locate with genes and change with allele-specific gene regulation.
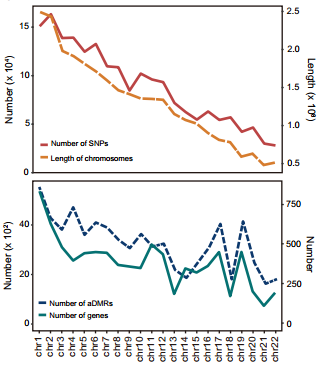
Figure 6: Allele-specific methylation regions profile
In this study, the researchers demonstrated that personal transcriptomes and personal DNA methylomes displayed distinct dynamics that were associated with different physiological conditions. Significant changes of the former often occurred during more dynamic and acute health conditions, whereas the PBMC DNA methylome profiles were linked to chronic events such as glucose level elevation.
In summary, ‘no single type of omics data is sufficient to predict precise human health states, but it may be possible to integrate multiple types of omics data to improve prediction power for a wide variety of disease types.’

Figure 7: Members of Dan Xie research team of the National Key Laboratory of Biotherapy (The second from left is Kailing Tu, the fifth from left is Lin Xia, and the sixth from left is Dr. Dan Xie.)
Corresponding author:
Dan Xie.Dr. Xie is a specially-appointed researcher and Ph.D. advisor of the State Key Laboratory of Biotherapy, Sichuan University and Deputy Executive Director, Center for Precision Medicine, West China Hospital, Sichuan University.He undertakes a number of basic research projects such as major research projects and surface projects of the National Natural Science Foundation of China. He has a long history of researching in the fields of gene regulation network, liquid biopsy and single-cell sequencing. He has contributed to nearly 20 SCI papers as the corresponding author and/or first author, including papers in Cell, Nature Biotechnology, Nature Medicine, Cell Research, and Genome Research. He also serves as a special reviewer for Nature Communications, PLOS One, and Cell Discovery.
